By Melissa Dossett & Monica Montgomery
Over the past few weeks, we have started working in the lab to go beyond the book-learning aspect of our research and delve further into the hands-on aspect of it. We were unsure about what to expect at first, as the labs we have done up until this point have been with TAs and professors carefully guiding us through the procedures and techniques. With the help of each other, Somer Smith (another undergraduate student), and the guidance of Elizabeth Latham, lab time has become very exciting because we have to think for ourselves and design research experiments.
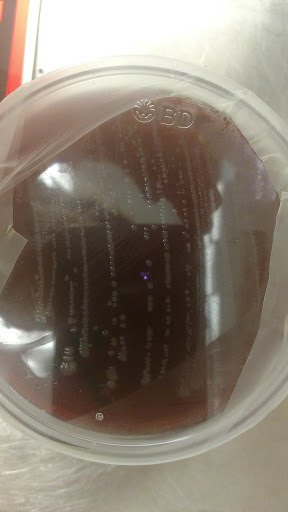
To begin our work in the laboratory, we plated different bacterial cultures that were growing on the moldy meat samples we had. It was not too pleasant of a sight, but it was fascinating to see the rapid growths of mold within the container! Next, we worked with a Qiagen DNA extraction kit which consisted of a great deal of centrifuging and pipetting. This step allowed us to isolate bacteria within the collected samples for further purification. PCR (Polymerase Chain reaction) was performed next. This process is used to amplify a small amount of DNA. We later attempted to utilize what we had obtained from the PCR to do a gel electrophoresis. Gel electrophoresis is a method used to separate mixtures of DNA or proteins according to molecular size. The molecules are separated by being pushed by an electrical field through a gel that contains small pores. The results from the gel electrophoresis were unsuccessful and could have been caused by one of three problems: 1) we did not get enough DNA amplification during PCR, 2) there was no DNA to begin with, or 3) there was improper pouring of the TAE buffer causing the ladder to contaminate other wells. Currently, we are still working to correct and troubleshoot problems for the gel electrophoresis.
Before summer break, the last experiment we left off on was using the DNA spectrophotometer to test for DNA purity within our samples. Unfortunately, the spectrophotometer results showed that our samples of DNA were impure. To fix this problem, we are going to fix errors in the plating and DNA extraction methods. There could possibly be cross contamination and incorrect handling of substances that we’ll get to the root in the next few days.
For the future, we are planning to perform tests on retrieved food samples from Elissa. Soon we will be designing experimental procedures for food collection and testing. We will further identify microbes, toxins, and their respective amounts in the lab and carry out a chemical analysis of suspected factors that may possibly be affecting the food. We will compile this data with our nutritional research to fully understand the nutrition and microbial intake of the average sailor from the beginning to end of a 3-month voyage. In addition, we will test promising microbes for medical and other beneficial uses. Given the unique preparation and storage of the food, we hope to discover probiotics that can improve people’s health today.
After troubleshooting errors within our previous experiments, we are looking forward to this next phase of research, as we are about to dive into our new laboratory objectives and hopefully discover the unknown!